In neuroimaging the typical analysis approach is a “mass univariate”, where a univariate model is independently fit to each voxel (volume element) in parallel. This approach can be applied to a large range of datasets including: functional MRI, anatomical MRI (with Voxel-Based Morphometry), PET, EEG and MEG data.
The three major software analysis package for neuroimaging: SPM (Statistical Parametric Mapping) and FSL (FMRIB Software Library) and AFNI (AFNI) provide implementations of mass univariate analyses. While there are many commonalities across software packages, there are also software-specific outputs that can be of interest for the end-user.
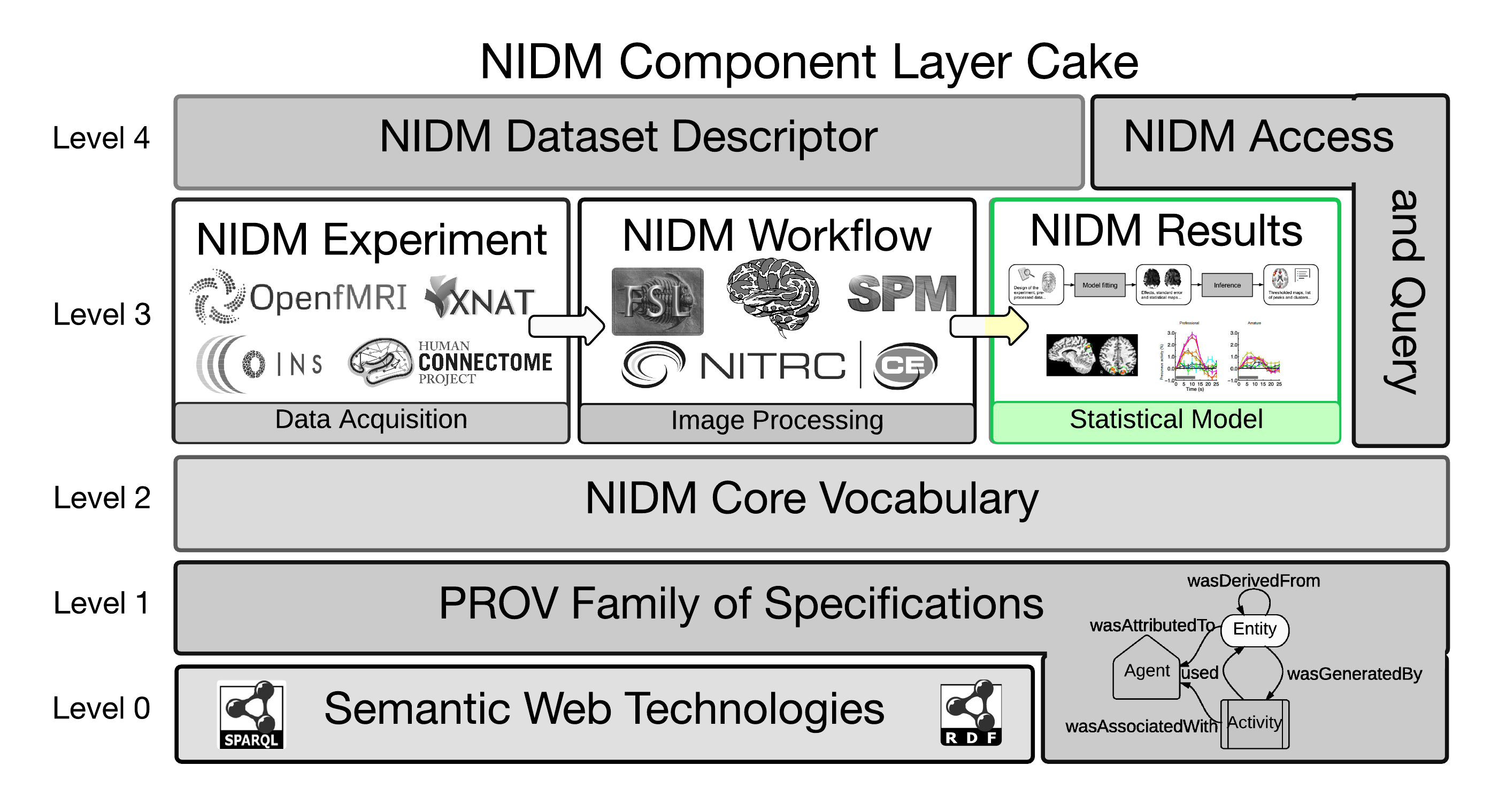
This document describes the encoding of results of a mass univariate neuroimaging analysis using the NIDM data model ([[!KEATORNI13]]). The goal of this specification is to provide a unified representation of neuroimaging results across analysis software. When a piece of information is only available in a specific software, software-specific extensions are provided.
NIDM-Results is a part of NIDM as depicted in .